Sample information |
||||||||||||||||||||||||||||||||||||||||
| Picture |

|
|||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location | ||||||||||||||||||||||||||||||||||||||||
| Collection date | 10/31/2024 | |||||||||||||||||||||||||||||||||||||||
| Captive / Cultivated? | Wild-caught | |||||||||||||||||||||||||||||||||||||||
| Group | Longwood University | |||||||||||||||||||||||||||||||||||||||
| Observations | Fall, 78 deg F |
|||||||||||||||||||||||||||||||||||||||
| Putative identification | Arthropoda Insecta Hymenoptera Scoliidae Scolia Scolia dubia | |||||||||||||||||||||||||||||||||||||||
Methods |
||||||||||||||||||||||||||||||||||||||||
| Extraction kit | Wizard SV (Promega) | |||||||||||||||||||||||||||||||||||||||
| DNA extraction location | Partial abdomen | |||||||||||||||||||||||||||||||||||||||
| Single or Duplex PCR | Single Reaction | |||||||||||||||||||||||||||||||||||||||
| Gel electrophoresis system | Standard electrophoresis system | |||||||||||||||||||||||||||||||||||||||
| Buffer | TAE | |||||||||||||||||||||||||||||||||||||||
| DNA stain | Ethidium Bromide | |||||||||||||||||||||||||||||||||||||||
| Gel images |
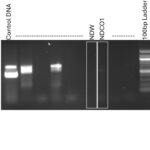
|
|||||||||||||||||||||||||||||||||||||||
| Protocol notes | In a 1.5 mL microfuge tube, the specimen and 275 µL of prepared Digestion Solution Master Mix, containing proteinase K, RNase A Solution, and 0.5 M EDTA. A pestle was used to physically grind the specimen and placed in a 55 ℃ water bath for 15 minutes for cell lysis. The sample was centrifuged for 60 seconds at 2000 x g and transferred to a new 1.5 mL microfuge tube with 250 µL of Wizard SV Lysis Buffer. The lysate sample was centrifuged for 3 minutes at 13,000 x g. The column was washed with 650 µL of Wizard SV Wash and centrifuged for 60 seconds at 13,000 x g. This wash process was repeated four times and 50 µL of 65 ℃ nuclease-free water was added. The eluted samples were stored at -20 ℃. Polymerase Chain Reaction The Arthropod PCR Cocktail was prepared in a 1.5 mL microcentrifuge tube with adequate measurements (Table 2), cortexed and centrifuged for proper mixing. Two 0.2 mL PCR tubes were tabled with control and extracted specimen DNA. In each 0.2 mL tube, 23 µL of the Arthropod PCR Cocktail was added and centrifuged. In the corresponding labeled PCR tubes, 2 µL of control and extracted specimen DNA was added. The samples were transferred to a BioRad Thermal Cycler and set for 2 minutes at 94°C for initial denaturation, 30 cycles at 30:45:60 second intervals at 94:49:72°C respectively, final extension was for 10 minutes at 72°C and held at 4°C. The 0.2 mL PCR tubes were stored in -20 ℃.
The 16s rRNA Wolbachia primer was prepared in a 1.5 mL PCR tube with the adequate measurements (Table 2). Two 0.2 mL PCR tubes were tabled with control and extracted specimen DNA. In each 0.2 mL tube, 23 µL of the Wolbachia primer mix was added and centrifuged. In the corresponding labeled PCR tubes, 2 µL of control and extracted specimen DNA was added. The samples were transferred to a BioRad Thermal Cycler and set for 2 minutes at 94°C for initial denaturation, 30 cycles at 30:45:60 second intervals at 94:55:72°C respectively, final extension was for 10 minutes at 72°C and held at 4°C. The 0.2 mL PCR tubes were stored in -20 ℃. Gel Electrophoresis A 1% agarose solution was prepared with 50 mL 1x TAE buffer and 0.5 g agarose microwaved. After the solution was clear, 5 µL of 10mg/mL ethidium bromide was added and then solidified in 30 minutes. The gel was covered 1 cm by 1x TAE Buffer prior to sample loading. el loading order is shown in Table 3.
The gel was run at 110 V for 30 minutes. DNA amplicon visualization is conducted in a UV SYNGENE Gel documentation system. System lighting is adjusted to capture the band sizes. The gel was disposed of in a biohazard waste container. PCR samples were sent to Applied Biosystems for Sanger sequencing. Bioinformatic Analysis Gel electrophoresis results were submitted to The Wolbachia Project database. Data input guidelines are provided by the database. Arthropod and Wolbachia PCR samples underwent Sanger sequencing at Applied Biosystems. Chromatogram data was received back in .ab1 format and examined in Benchling software. Sequence realignment trimmed reads with a QS value under 40 on the 5’ and 3’ end. A FASTA file was created following realignment and was imported to Primer-BLAST (National Library of Medicine 2024). Core_nts database was used in Primer-BLAST. The discrepancy of BLAST genetic identification and Seek identification will be discussed later. Phylogenetic analysis was conducted in NGPhylogeny.fr (Lemoine et al. 2019) and sample CO1 gene sequences were provided for 11 arthropod species. |
|||||||||||||||||||||||||||||||||||||||
Results |
||||||||||||||||||||||||||||||||||||||||
| Wolbachia presence | No | |||||||||||||||||||||||||||||||||||||||
| Confidence level | Low | |||||||||||||||||||||||||||||||||||||||
| Explanation of confidence level | No band showed up in Wolbachia but controls worked. Wolbachia positive worked but the sample did not work. Organism did not ID correctly either. Amplification of a wrong organism results in low confidence. |
|||||||||||||||||||||||||||||||||||||||
| Wolbachia 16S sequence | Download FASTA
|
|||||||||||||||||||||||||||||||||||||||
| Arthropod COI sequence | Download AB1
|
|||||||||||||||||||||||||||||||||||||||
| Summary | The Scolia dubia was found to be negative for Wolbachia. | |||||||||||||||||||||||||||||||||||||||
 European Wasp
European Wasp Small Honey Ant
Small Honey Ant 7A
7A Millipede 7F
Millipede 7F