Sample information |
|
| Picture |

|
|---|---|
| Location | |
| Collection date | 11/13/2024 |
| Captive / Cultivated? | Wild-caught |
| Group | Hawaii Community College |
| Observations |
|
| Putative identification | Arthropoda Insecta Diptera |
Methods |
|
| Extraction kit | pcr |
| DNA extraction location | Whole arthropod |
| Single or Duplex PCR | Single Reaction |
| Gel electrophoresis system | Edvotek Gel Electrophoresis |
| Buffer | TBE |
| DNA stain | SYBR Safe |
| Gel images |
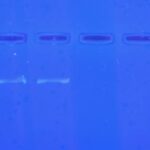
|
| Protocol notes | I obtained my arthropod samples via an apple cider vinegar trap. I believe the flies I collected are fruit flies, but not necessarily a drosophilid. To prepare the DNA for PCR, both flies were ground in separate vials 100 µL of cell lysis buffer, and then another 100 µL was added. These vials were heated at 95oC for five minutes and after being cooled back down were vortexed and then centrifuged for two minutes at the highest speed. 150 µL of the supernatant from each tube was transferred into a new tube with 150 µL of prepared cold isopropanol. These were then vortexed and centrifuged for five minutes. As much supernatant as possible from each tube was transferred to a waste cup, and they were allowed to dry for a minute. 100 µL of 70% ethanol was added to each tube, they were vortexed and centrifuged for a minute, and then the supernatant was poured out and the tubes were allowed to dry for 10 minutes. 50 µL of DNA elution buffer was added to each tube, and they were incubated in a 65OC bath until the PCR reaction. The professor completed the PCR reaction, so my next tasks were to prepare the agarose gel. With a lab partner I prepared a double 6 well comb, by first adding 1 g of agarose gel to 50 mL of 1x TBE solution. We heated this in a microwave until the solution appeared clear, after cooling down to a safe amount we added 5 µL SYBR safe DNA dye and poured the solution in to the tray. After the gel set and the combes and rubber caps were removed, it was covered entirely in TBE buffer. I used parafilm to set up 5 drops of 2 µL 6X DNA loading dye. The first drop was mixed with a 10 µL of DNA ladder via a micropipette and loaded into the gel. The rest mixed with 10 µL DNA samples from my the PCR reactions of my arthropods. The actual electrophoresis and photography from the transilluminator were also done by the professor. |
Results |
|
| Wolbachia presence | No |
| Confidence level | High |
| Explanation of confidence level | The positive control done by the professor clearly showed Wolbachia lines below the DNA ladder, not seen from either of my samples. |
| Wolbachia 16S sequence | |
| Arthropod COI sequence |
|
| Summary | The Diptera was found to be negative for Wolbachia. |
 Nothing out of the ordinary.
Nothing out of the ordinary. European Wasp
European Wasp Small Honey Ant
Small Honey Ant 7A
7A Millipede 7F
Millipede 7F